RS/tRNA Foundational Publication Support
Erdmann, Ines, Kathrin Marter, Oliver Kobler, Sven Niehues, Julia Abele, Anke Müller, Julia Bussmann, et al. (2015) 2015. “Cell-Selective Labelling Of Proteomes In Drosophila Melanogaster.”. Nature Communications 6: 7521. doi:10.1038/ncomms8521.
RS/tRNA Usage Publications
Niehues, Sven, Julia Bussmann, Georg Steffes, Ines Erdmann, Caroline Köhrer, Litao Sun, Marina Wagner, et al. (2015) 2015. “Impaired Protein Translation In Drosophila Models For Charcot-Marie-Tooth Neuropathy Caused By Mutant Trna Synthetases.”. Nature Communications 6: 7520. doi:10.1038/ncomms8520.
RS/tRNA Pair Development Year
2015
ncAA(s) Incorporated
Azidonorleucine
ncAA Structure (png, jpg, jpeg)
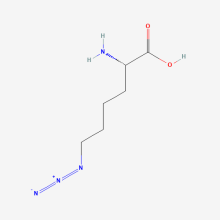
ncAA Utility
Biorthogonal Ligation, Click Chemistry labeling with fluorescence or affinity labels. Specifically used for cell-specific proteomics studies using "bioorthogonal noncanonical amino acid tagging" (BONCAT) proteomics. Contrasts with Azidohomoalanine which does not allow cell-specific studies.
RS Organism of Origin
Parent RS
RS Mutations
L262G
tRNA Organism of Origin
Parent tRNA
tRNA Anticodon
CAU
Multiple tRNAs?
may also be able to pair with other eukaryotic endogenous Met tRNAs
RS/tRNA Availability
n/a
Used in what cell line?
RS/tRNA Additional Notes
Was used to label proteins in live flies for proteomics studies. Both this drosophila RS and the mouse RS with equivalent mutations (L274G MmMetRS) worked in live flies, so the authors used the general designator MetRS(LtoG) to cover both versions. The fly version was slightly less disruptive to fly development.
