RS/tRNA Foundational Publication Support
Miyake-Stoner, Shigeki J, Andrew M Miller, Jared T Hammill, Jennifer C Peeler, Kenneth R Hess, Ryan A Mehl, and Scott H Brewer. (2009) 2009. “Probing Protein Folding Using Site-Specifically Encoded Unnatural Amino Acids As Fret Donors With Tryptophan.”. Biochemistry 48 (25): 5953-62. doi:10.1021/bi900426d.
Peeler, Jennifer C, and Ryan A Mehl. (2012) 2012. “Site-Specific Incorporation Of Unnatural Amino Acids As Probes For Protein Conformational Changes.”. Methods In Molecular Biology (Clifton, N.j.) 794: 125-34. doi:10.1007/978-1-61779-331-8_8.
Beyer, Jenna N, Parisa Hosseinzadeh, Ilana Gottfried-Lee, Elise M Van Fossen, Phillip Zhu, Riley M Bednar, Andrew Karplus, Ryan A Mehl, and Richard B Cooley. (2020) 2020. “Overcoming Near-Cognate Suppression In A Release Factor 1-Deficient Host With An Improved Nitro-Tyrosine Trna Synthetase.”. Journal Of Molecular Biology 432 (16): 4690-4704. doi:10.1016/j.jmb.2020.06.014.
RS/tRNA Protocols and Structural Information
Peeler, Jennifer C, and Ryan A Mehl. (2012) 2012. “Site-Specific Incorporation Of Unnatural Amino Acids As Probes For Protein Conformational Changes.”. Methods In Molecular Biology (Clifton, N.j.) 794: 125-34. doi:10.1007/978-1-61779-331-8_8.
Cooley, Richard B, and Holger Sondermann. (2017) 2017. “Probing Protein-Protein Interactions With Genetically Encoded Photoactivatable Cross-Linkers.”. Methods In Molecular Biology (Clifton, N.j.) 1657: 331-345. doi:10.1007/978-1-4939-7240-1_26.
RS/tRNA Usage Publications
Bednar, Riley M, Subhashis Jana, Sahiti Kuppa, Rachel Franklin, Joseph Beckman, Edwin Antony, Richard B Cooley, and Ryan A Mehl. (2021) 2021. “Genetic Incorporation Of Two Mutually Orthogonal Bioorthogonal Amino Acids That Enable Efficient Protein Dual-Labeling In Cells.”. Acs Chemical Biology 16 (11): 2612-2622. doi:10.1021/acschembio.1c00649.
Chatterjee, Debashree, Richard B Cooley, Chelsea Boyd, Ryan A Mehl, George O'Toole, and Holger Sondermann. (2014) 2014. “Mechanistic Insight Into The Conserved Allosteric Regulation Of Periplasmic Proteolysis By The Signaling Molecule Cyclic-Di-Gmp.”. Elife 3: e03650. doi:10.7554/eLife.03650.
Cooley, Richard B, John O'Donnell, and Holger Sondermann. (2016) 2016. “Coincidence Detection And Bi-Directional Transmembrane Signaling Control A Bacterial Second Messenger Receptor.”. Elife 5. doi:10.7554/eLife.21848.
Gwyther, Rebecca, Sébastien Côté, Chang-Seuk Lee, Haosen Miao, Krithika Ramakrishnan, Matteo Palma, and Dafydd Jones. (2024) 2024. “Optimising Cnt-Fet Biosensor Design Through Modelling Of Biomolecular Electrostatic Gating And Its Application To Β-Lactamase Detection.”. Nature Communications 15 (1): 7482. doi:10.1038/s41467-024-51325-6.
RS/tRNA Pair Development Year
2009
ncAA(s) Incorporated
p-cyano-L-phenylalanine
ncAA Structure (png, jpg, jpeg)
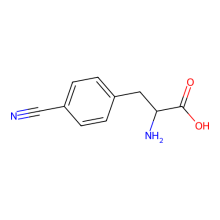
ncAA Utility
FRET Probe (with Trp) and infrared probe
p-ethynyl-L-phenylalanine
ncAA Structure (png, jpg, jpeg)
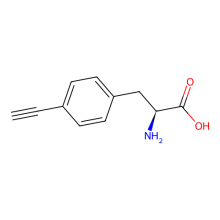
ncAA Utility
FRET Probe
p-azido-L-phenylalanine (pAzF)
ncAA Structure (png, jpg, jpeg)
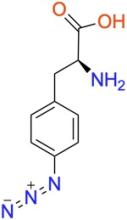
ncAA Utility
Used as a photocrosslinker, allowing for crosslinking and bioorthogonal click-chemistry ligation of proteins via strain-promoted cycloadditions (SPAAC) with suitably functionalized molecules.
RS Organism of Origin
Parent RS
RS Mutations
Y32L
L65V
F108W
Q109M
D158G
I159P
L65V
F108W
Q109M
D158G
I159P
tRNA Organism of Origin
Parent tRNA
tRNA Anticodon
CUA
Other tRNA Mutations
C17A
U17aG
U20C
G37A
U47G
U17aG
U20C
G37A
U47G
Multiple tRNAs?
N/a
RS/tRNA Availability
On pDule plasmid as Addgene #85494
On pDule2 plasmid as Addgene #85495
On pEVOL plasmid as Addgene #164579 designed for use in dual encoding applications involving pAzido-Phe (from 2021 Bednar et al application paper)
On pDule2 plasmid as Addgene #85495
On pEVOL plasmid as Addgene #164579 designed for use in dual encoding applications involving pAzido-Phe (from 2021 Bednar et al application paper)
Used in what cell line?
RS/tRNA Additional Notes
"p(sp)Phe RS" (i.e. an RS incorporating multiple "sp" hybridized para-substituents on Phe) selections led to an RS incorporating both p-cyano-Phe (pCNF) and pENPhe. With 1 mM L-pCNF generated T4 lysozyme(153) at ~7.5 mg/L (~25% effciency). With 1 mM racemic pENPhe, yielded 23 mg/L protein (~75% efficiency). Full fidelity was confirmed by MS.
Shown in 2012 foundational (and methods) paper to also incorporate p-azidoPhe (pAzF) similarly to pENPhe.
2019 foundational paper showed also produces full-fidelity incorporation of pAzF into sfGFP(150) in the truncation free B95 RF1- E coli strain.
2017 methods paper includes pAzF incorporation and photocrosslinking protocols.
Shown in 2012 foundational (and methods) paper to also incorporate p-azidoPhe (pAzF) similarly to pENPhe.
2019 foundational paper showed also produces full-fidelity incorporation of pAzF into sfGFP(150) in the truncation free B95 RF1- E coli strain.
2017 methods paper includes pAzF incorporation and photocrosslinking protocols.
