RS/tRNA Foundational Publication Support
Amiram, Miriam, Adrian D. Haimovich, Chenguang Fan, Yane-Shih Wang, Hans-Rudolf Aerni, Ioanna Ntai, Daniel W. Moonan, et al. (dec) 2015. “Evolution Of Translation Machinery In Recoded Bacteria Enables Multi-Site Incorporation Of Nonstandard Amino Acids”. Nature Biotechnology 33: 1272-1279. doi:10.1038/nbt.3372.
Wang, Lei, Zhiwen Zhang, Ansgar Brock, and Peter G Schultz. (2003) 2003. “Addition Of The Keto Functional Group To The Genetic Code Of Escherichia Coli.”. Proceedings Of The National Academy Of Sciences Of The United States Of America 100 (1): 56-61.
Young, Travis S, Insha Ahmad, Jun A Yin, and Peter G Schultz. (2010) 2010. “An Enhanced System For Unnatural Amino Acid Mutagenesis In E. Coli.”. Journal Of Molecular Biology 395 (2): 361-74. doi:10.1016/j.jmb.2009.10.030.
Young, Douglas D, Travis S Young, Michael Jahnz, Insha Ahmad, Glen Spraggon, and Peter G Schultz. (2011) 2011. “An Evolved Aminoacyl-Trna Synthetase With Atypical Polysubstrate Specificity.”. Biochemistry 50 (11): 1894-900. doi:10.1021/bi101929e.
RS/tRNA Usage Publications
Bak, Mijeong, Junyong Park, Kiyoon Min, Jinhwan Cho, Jihyoun Seong, Young S Hahn, Giyoong Tae, and Inchan Kwon. (2020) 2020. “Recombinant Peptide Production Platform Coupled With Site-Specific Albumin Conjugation Enables A Convenient Production Of Long-Acting Therapeutic Peptide.”. Pharmaceutics 12 (4). doi:10.3390/pharmaceutics12040364.
Park, Junyong, Mijeong Bak, Kiyoon Min, Hyun-Woo Kim, Jeong-Haeng Cho, Giyoong Tae, and Inchan Kwon. (2021) 2021. “Effect Of C-Terminus Conjugation Via Different Conjugation Chemistries On In Vivo Activity Of Albumin-Conjugated Recombinant Glp-1.”. Pharmaceutics 13 (2). doi:10.3390/pharmaceutics13020263.
RS/tRNA Pair Development Year
2015
ncAA(s) Incorporated
p-azido-L-phenylalanine (pAzF)
ncAA Structure (png, jpg, jpeg)
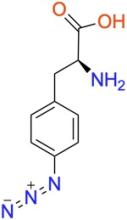
ncAA Utility
Used as a photocrosslinker, allowing for crosslinking and bioorthogonal click-chemistry ligation of proteins via strain-promoted cycloadditions (SPAAC) with suitably functionalized molecules.
RS Organism of Origin
Parent RS
RS Mutations
Y32L
D158V
I159M
L162D
A167Y
R257G
D158V
I159M
L162D
A167Y
R257G
tRNA Organism of Origin
Parent tRNA
tRNA Anticodon
CUA
Other tRNA Mutations
C17A
U17aG
U20C
G37A
U47G
U17aG
U20C
G37A
U47G
RS/tRNA Availability
Addgene plasmid #73547
Used in what cell line?
RS/tRNA Additional Notes
This RS/tRNA pair was evolved in the C321.ΔA E. coli strain from the AcetylPhe RS (pAcF-RS) originally reported in in 2002 foundational paper, used as a model RS in the 2010 foundational paper and characterized for permissivity in the 2011 foundational paper. In 2015 further residues were allowed to evolve to optimize p-azido-Phe and tRNA interactions and improve expression of multiple ncAA containing proteins using low RS levels. This evolved RS showed ~6-fold higher expression of pAzF in GFP-3sites compared to the progenitor enzyme (pAcFRS) using a chromosome incorporated RS. Figure 4a of foundational paper shows that when included as multicopy plasmid it still highly outperforms the starting RS for incorporating 30 ncAAs in a construct, but only outperforms the original unevolved RS form by ~2-fold when expressing GFP with 3 ncAAs, and that along with the improved expression is a much lower fidelity (higher protein production in the absence of ncAA). Note that the "pAzRS" in this figure is the pCNFRS shown in the 2011 foundational paper to work very well for pAzF. This RS was also shown to be quite specific for pAzF, and among the RSs evolved in this work, it seems the best one for pAzF incorporation due to its higher fidelity compared with pAzFRS.2.t1.
